Single-cell Colorectal Cancer Atlas
Single-cell integration and multi-modal profiling reveals phenotypes and spatial organization of neutrophils in colorectal cancer
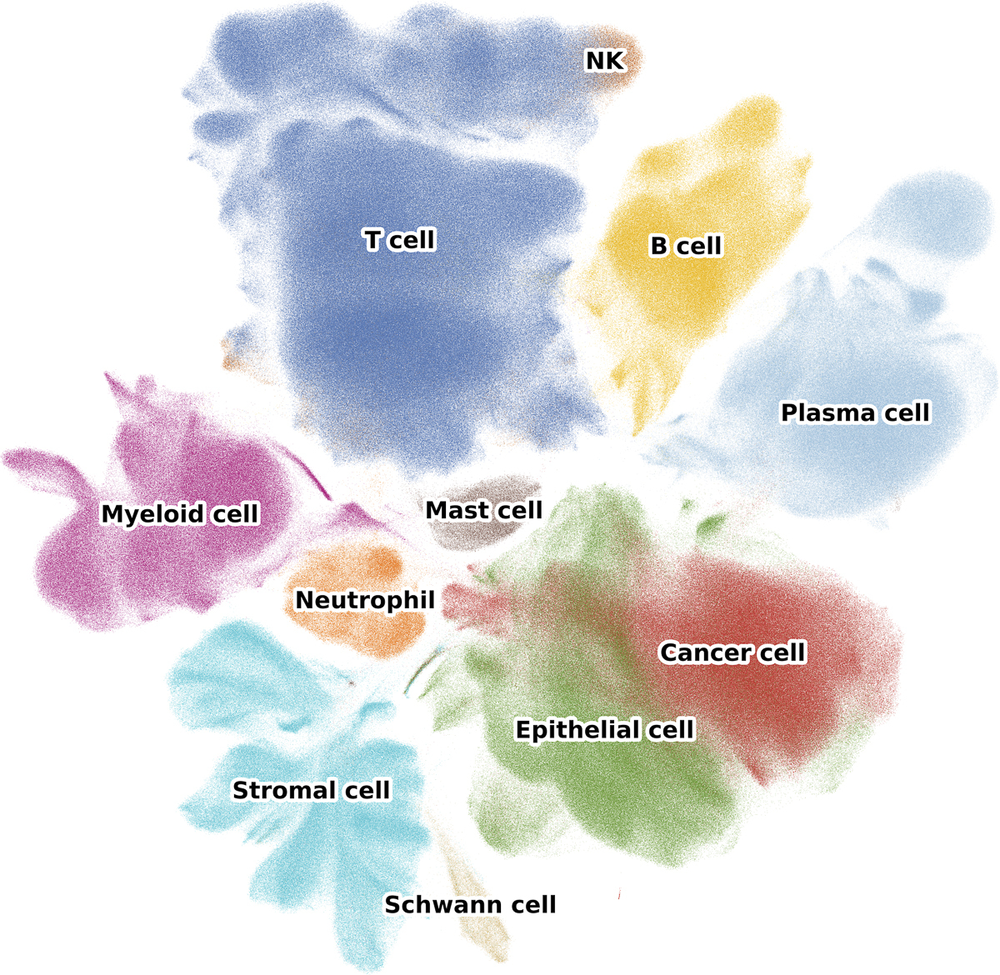
The single cell colorectal cancer atlas is a resource integrating more than 4.27 million cells from 650 patients across 49 studies (77 datasets) representing 7 billion expression values. These samples encompass the full spectrum of disease progression, from normal colon to polyps, primary tumors, and metastases, covering both early and advanced stages of CRC. Additionally, to address the limited availability of neutrophil single-cell data, we supplemented the atlas by analyzing samples from 12 CRC patients using a platform that captures cells with very low transcript counts. We present a high-resolution view of CRC with 62 major cell types or states, revealing different cell-type composition patterns in CRC subtypes.
Browse or get the full CRC atlas
Browse and interactively explore the full atlas on our local cellxgene instance.
If you are interested in running your
own analyses on the full atlas, you can download the
h5ad file with standardized metadata.
CZI cell-x-gene portal
The core-atlas will become available on the CZI
cell-x-gene soon. There, you will be able to
explore the datasets interactively in the browser and
download them as h5ad (scverse) or rds (Seurat) files with standardized
metadata for further analysis.
Browse code
All code needed to reproduce the study is wrapped into a nextflow workflow and publicy available from GitHub.
Download additional data
In addition to the h5ad objects mentioned
above, we provide preprocessed input data, intermediate
results and final results on zenodo.
Spatial transcriptomics (Xenium) and Imaging Mass Cytometry (IMC) data may be downloaded from BioImage Archive.
Contact
For reproducibility issues or any other requests regarding single-cell data analysis, please use the issue tracker. For anything else, you can reach out to the corresponding author(s) as indicated in the manuscript.